Services
NGS
Amplicon Metagenomic Sequencing
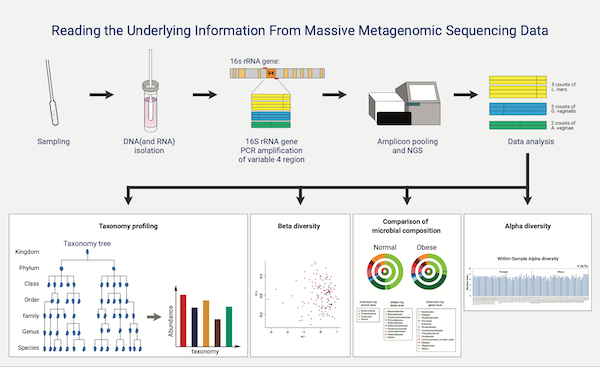
Amplicon metagenomic sequencing is an efficient and economical sequencing method for identifying and comparing bacteria, fungi, and other microorganisms. NGS-based analysis allows for taxonomic comparisons of complex microbiomes or cluster diversities within an environment that are difficult or impossible to culture and provides visual and statistical indicators to aid researchers' understanding. If Amplicon sequencing is not using a universal primer set, sufficient consultation is required for experimentation and analysis.
Application
- Taxonomic profiling
- Comparision analysis of microbial community
- Diversity statistics (alpha-, beta- diversity)
Sample/Laboratory Information
| Sample Requirement | gDNA > 100ng (minimum 10ng) 20ng/µl DIN > 6 |
|---|---|
| Library Kit | Illumina |
| Sequencing Platform | Illumina MiSeq |
| Recommended Sequencing Depth | Up to 50,000 reads per sample |
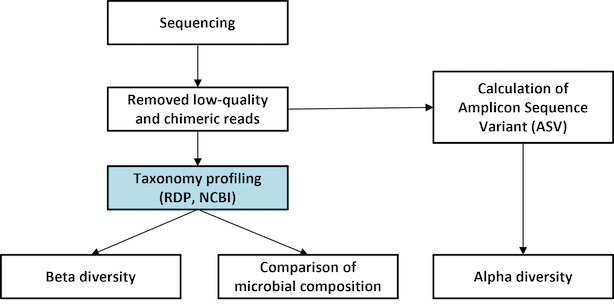
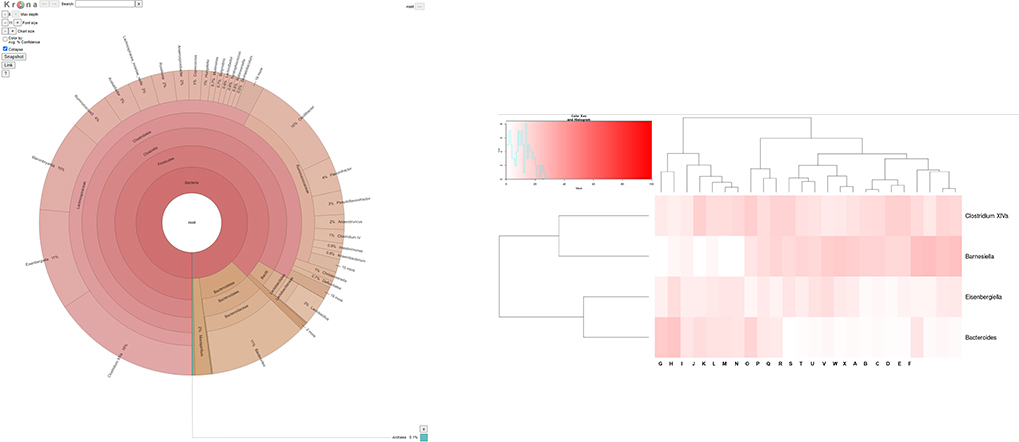
Bioinformatics
- Workflow
-
-
Standard Analysis
Pick De novo OTUs
Microbioal taxonomic Profiling
Diversity statistics(alpha-, beta- diversity)
Reference
[1] Lim, M. Y. et al., (2020). Changes in microbiome and metabolomic profiles of fecal samples stored with stabilizing solution at room temperature: a pilot study. Scientific reports, 10(1), 1789.
[2] Gunathilake, M. N. et al., (2019). Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Scientific reports, 9(1), 13589.
[3] Hu, H. J. et al., (2015). Obesity Alters the Microbial Community Profile in Korean Adolescents. PloS one, 10(7), e0134333.
[4] Saha, S. et al., (2019). Microbial acclimatization to lipidic-waste facilitates the efficacy of acidogenic fermentation. Chemical Engineering Journal, 358, 188–196. https://doi.org/10.1016/j.cej.2018.09.220
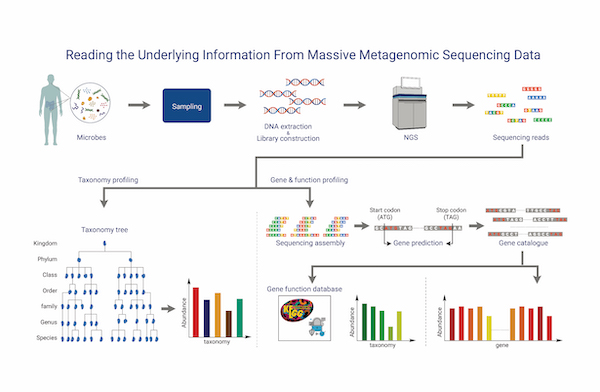
Shotgun Metagenomic Sequencing
Shotgun metagenomic sequencing based on NGS can comprehensively detect genes from all organisms, including uncultivable microorganisms or those in complex environments, providing information on microbial diversity and functional characteristics to aid in understanding relationships between microorganisms and their environments. It provides contig-level assembly of all organisms present in the sample, and gene annotation is performed by predicting CDS regions on the genome and utilizing databases such as UniProt, RefSeq, and Pfam.
Application
- Identification of microbial community diversity in the environmnet
- Metagenome assembled genomes
Sample/Laboratory Information
| Sample Requirement | gDNA > 10µg (minimum 5µg), 100ng/µl, DIN > 8 |
|---|---|
| Library Kit | Illumina |
| Sequencing Platform | Illumina NovaSeq6000, PacBio Sequel Ⅱ |
| Recommended Sequencing Reads | 70 million read pairs per sample |
Bioinformatics
- Workflow
-
-
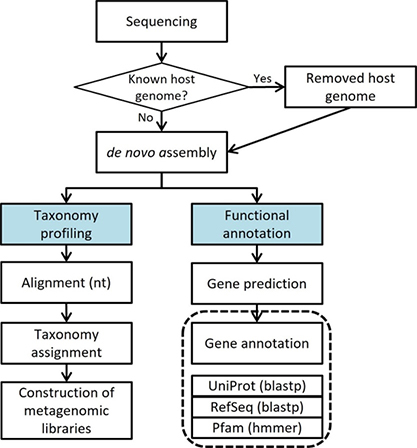
Standard Analysis
Remove host sequences
Metagenome assembled genomes
Prediction genomic region
Gene annotation
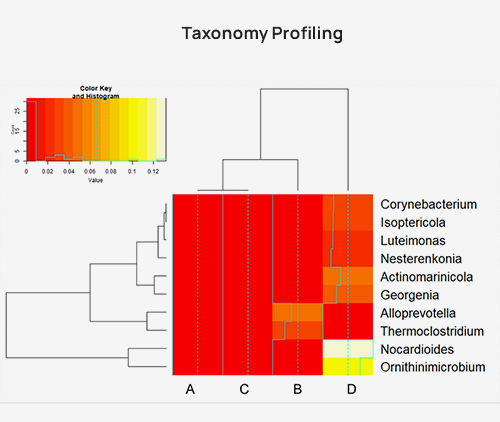
Taxonomy assign
-
Advanced Analysis
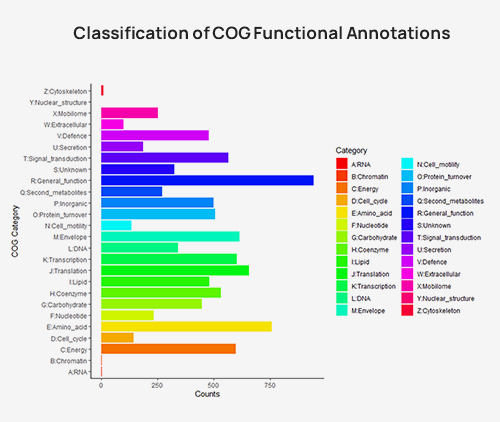
Classification of COG functional annotations
No content
No content
No content
No content
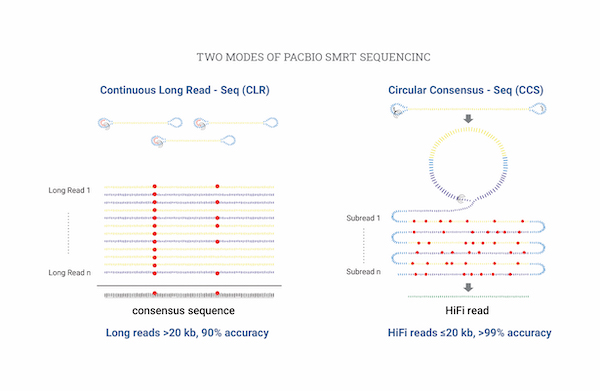
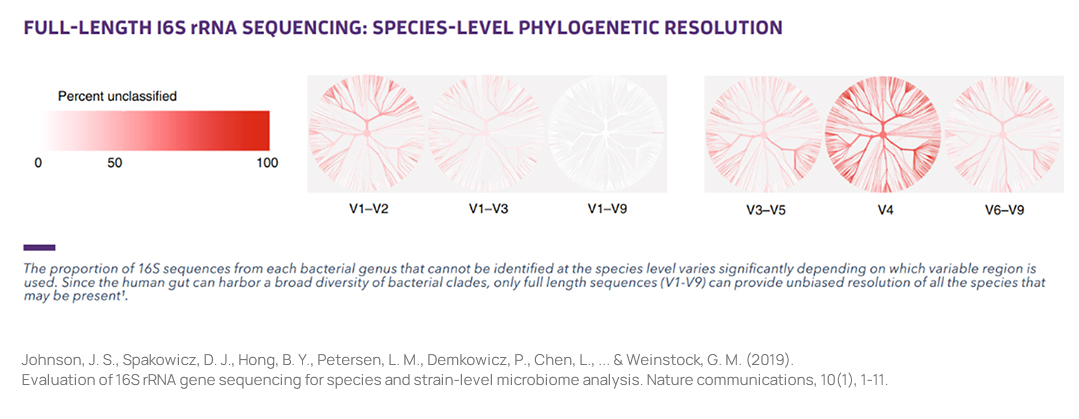
Full Length 16S rRNASequencing
NGS-based microbiome research has the challenge of misclassifying at the species level due to the short read length, despite the advantage of low cost. PacBio's Single Molecule, Real-Time (SMRT) sequencing can decode approximately 1.5 Kbp in the 16S rRNA and 18S rRNA regions, providing highly accurate and high-resolution profiling results at the species level.
Application
- Microbial taxonomic profiling
- Full length 16S rRNA genomic analysis
Sample/Laboratory Information
| Sample Requirement | gDNA > 1µg (minimum 5µg), 100ng/µl, DIN > 8 |
|---|---|
| Library Kit | PacBio |
| Sequencing Platform | PacBio Sequel Ⅱ |
| Recommended Sequencing Depth | - |
Bioinformatics
- Workflow
-