R&D
DeepOmicsTM
AI-based customized analysis platform
Theragen Bio has its own DEEPOMICS® platform, a unique genome analysis method, by combining more than a decade of
genome analysis experiences and research capabilities with artificial intelligence research capabilities.
TheDEEPOMICS® platform is an artificial intelligence platform that automatically executes the entire process of
disease classification (Molecular subtype classification in cancer), subtype specific biomarker discovery, therapeutic target discovery,
in silico drug screening, and in silico de novo drug design.
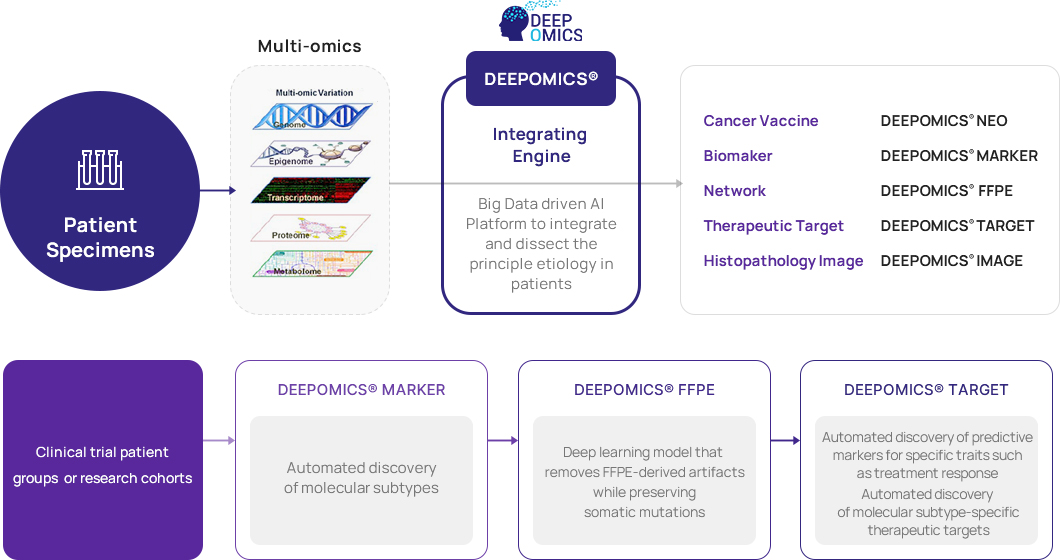
DEEPOMICS® NEO
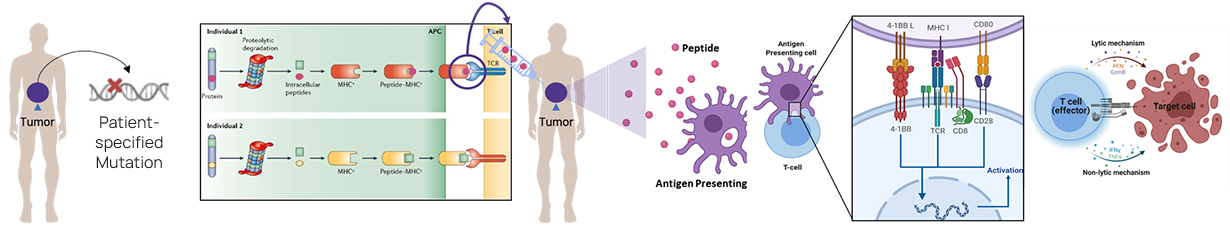
DEEPOMICS® NEO identifies somatic cell mutations present in cancer tissues of patient
and predicts neoantigens for individual MHCs.
- Patient-specific
mutation identification

- Prediction of
neopeitopes

- Neoepitope
Vaccivation

- Immune activation
by neoepitopes

- Cancer cell
death

Unlike the existing methodology, which only considered the combination of MHC and new antigen,DEEPOMICS® NEO is a deep learning model that considers
East Asian-specific rare MHC as well asthe activity of T cells, showing superior performance compared to other methodologies.
DEEPOMICS® NEO Achievement
Innovative technology that improved immunogenicity
predictability to the highest level in the world
It is a groundbreaking artifical intelligence technology that predicts HLA restricted neoepitopes among mutations identified through sequencing cancer tissue. Theragen Bio achieved three registered patents.

HLA ClassⅠ
-
Patent 1. HLA ClassⅠ
A model that learned through deep learning the ‘binding’ between HLA and peptide, which the existing algorithms focused on, and the ‘immunogenicity’ that stimulates T cells after binding
Both aspects of "Immunogenicity" learned through deep learning
(① Immunogenicity based on amino acid sequence of the peptides
② Immunogenicity based on T-cell recognition of peptide-MHC complex)

HLA ClassⅡ
-
Patent 2. HLA ClassⅡ
Predicting a neoantigen that binds to HLA Class II, where a combination of alpha and beta chains occurs, and deriving a synthetic long peptide neoantigen vaccine candidates reflecting both Class I and Class II
Inducing the activity of CD4+ T cells as well as CD8+ T cells in patients results in superior therapeutic efficacy

Rare HLA
Solution-
Patent 3. Rare HLA, Provide solutions for rate HLA types and varying length of neoepitopes
Applying modeling to overcome the lack of data for rare HLA alleles
Overcoming the problem of unstable predictions for East Asian HLA types
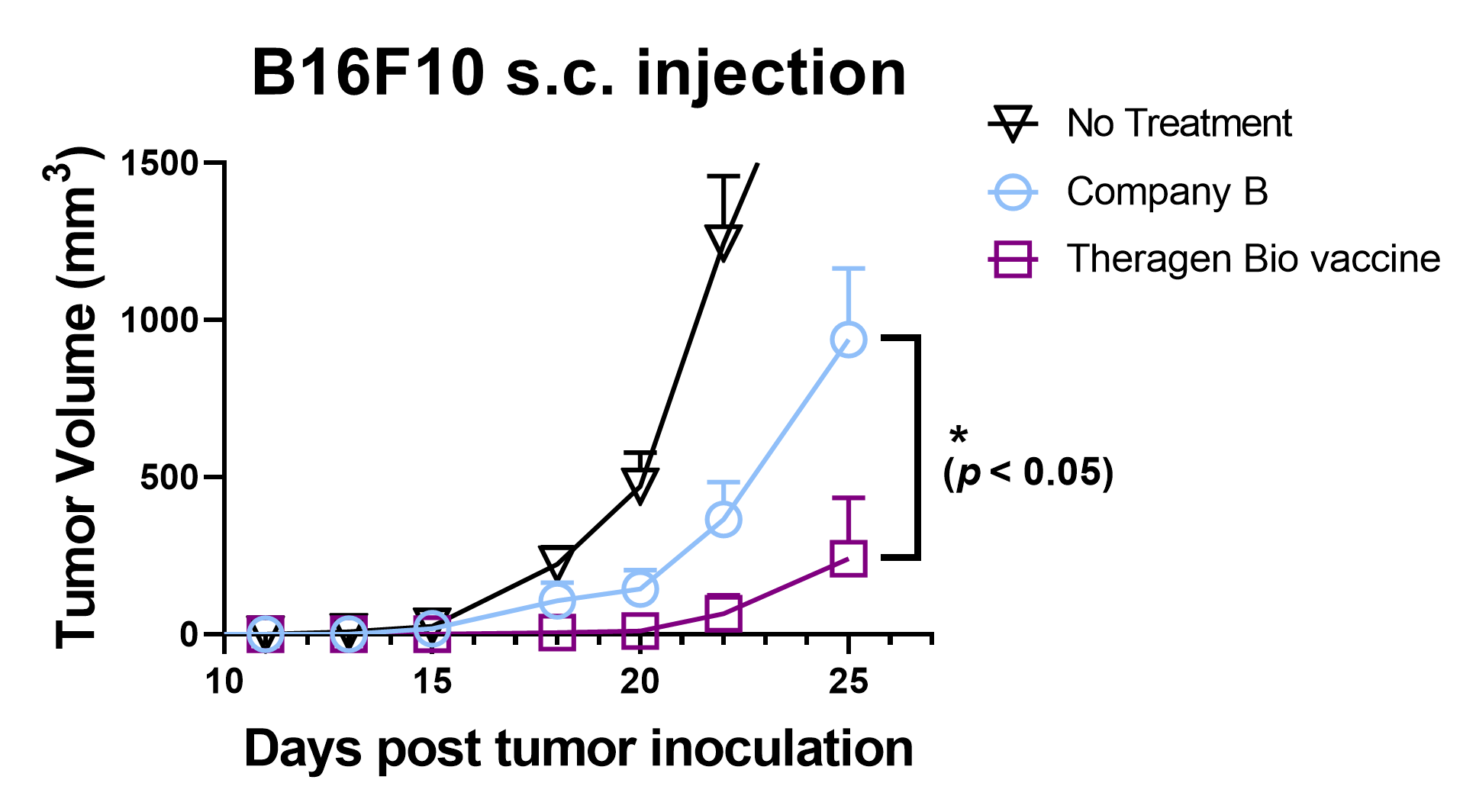
Superior efficacy of neoepitope vaccine designed with DEEPOMICS® NEO
mRNA vaccine designed with DEEPOMICS® NEO showed superior efficacy over vaccine design by the global leader in B6F10 melanoma tumor model.
DEEPOMICS® MARKER
DEEPOMICS® MARKER is a platform that automatically identifies disease subtypes and develops subtype classifiers.
Patients-classified biomarker
- Patient Classification
- Biomarker
- CDx
- Classification of molecular subtypes
- Automatic classification of molecular subtypes with different characteristics for the target disease.
- Disease
transcriptomics - Major characteristics derived from transcriptional data of the target disease without normal control group.
- Characteristics of molecular subtypes
- Discovery of specific major genes and signatures for each molecular subtype based on the disease transcriptomics
- Deep learning-based
classification model - Suggestion of deep learning classification model based on the discovered major signature
Compared to the existing molecular subtype classification method, DEEPOMICS® MARKER provides finer classification with superior clinical applicability, which ultimately links to DEEPOMICS® NETWORK and DEEPOMICS® TARGET for automatic suggestion for targeted therapeutics.
Application
- Patient classification of drug responses
- Suggestion of major signatures and genes vaild for drug responses
Differentiation of the method
for input processing
DEEPOMICS® MARKER utilizes Theragen Bio-specific data preprocessing that is completely different from the existing RNA-seq analysis (Patient No. 10-2385483). When cancer cell lines are analyzed using exisitng methods, many of them fail to map to their site of origin. When the same data are analyzed using DEEPOMICS® MARKER, mapping to the site of origin is much improved - suggesting the superiority of our analysis platform.
DEEPOMICS® FFPE
Deep Neural Network based on AI Model

-
A deep learning model that maximizes the preservation of somatic mutations while minimizing FFPE-derived Artifact.
* What is FFPE(Formalin-fixed, paraffin-embedded)?FFPE, which stands for formalin-fixed paraffin-embedded tissue, is a widely known method for preserving many cancer tissue samples by stabilizing proteins, nucleic acids, and overall structure. It provides important information for disease research, but during the process of fixing and processing the tissue, unnecessary nucleic acid mutations are induced, and RNA nucleic acids are fragmented. As a result, analysis becomes difficult and challenging, but FFPE is mainly used for companion diagnostic tests for targeted therapy.
DEEPOMICS® FFPE
Deep Neural Network based on AI Model
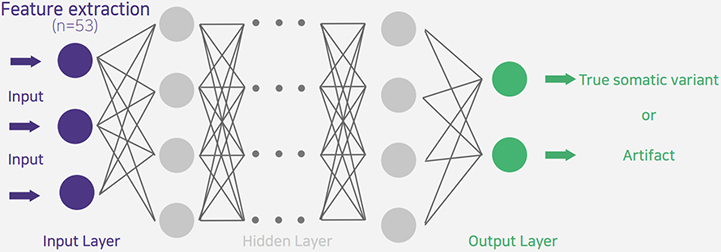
 [Figure1] shows an example of an FFPE sample
[Figure1] shows an example of an FFPE sample
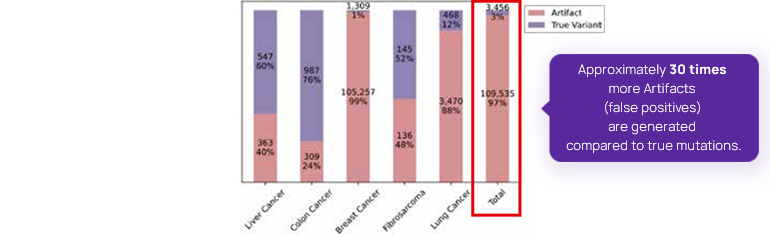 [Figure2] shows the Artifacts by cancer type
[Figure2] shows the Artifacts by cancer type
Far better technology than conventional technology
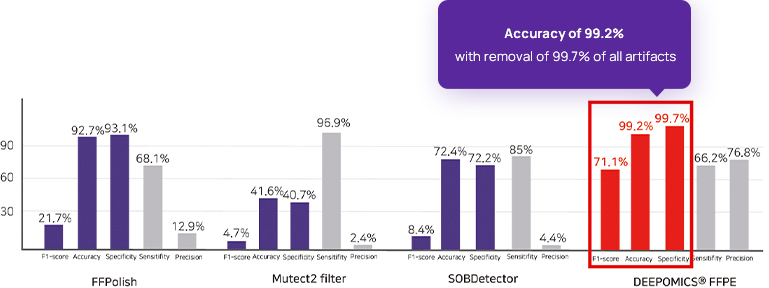
DEEPOMICS® FFPE
Workflow
-
Fastq files
-
1

Quality Control
FastQC -
2

Align to Reference
BWA -
3

Local realignment
GATK
Base quality recalibration -
4

Variant Calling
MuTect2
(SNPs and InDels) -
SnpEff annotation
-

DEEPOMICS® FFPE




