Services
Bioinformatics
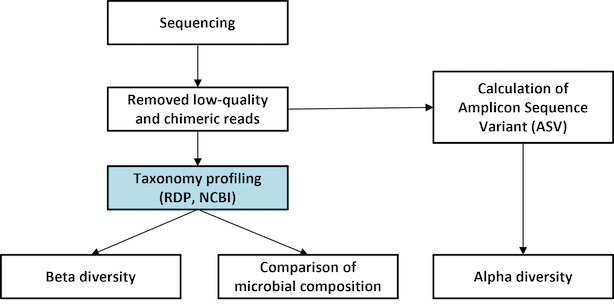
Amplicon Metagenomic Sequencing
- Workflow
-
Bioinformatics
-
Standard Analysis
Pick de novo OTUs
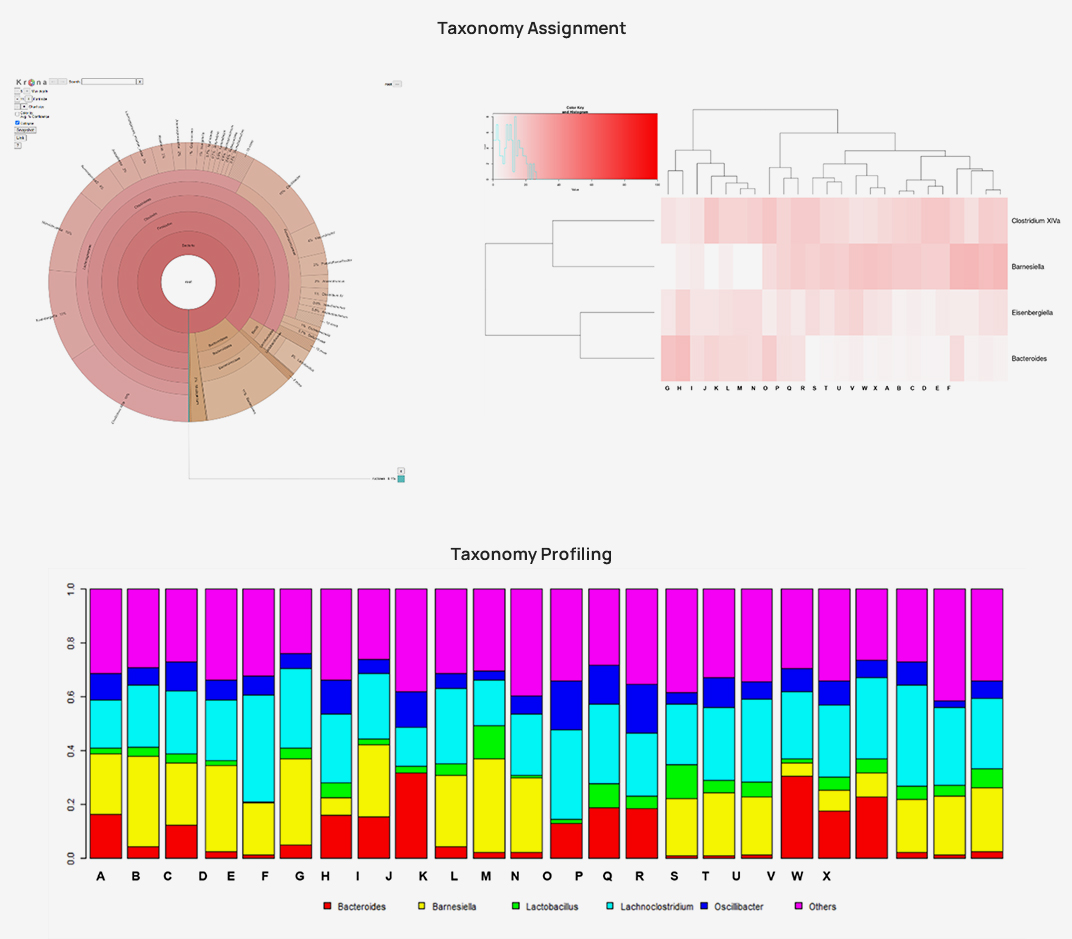
Taxonomic assignment
Diversity statistics (alpha-, beta- diversity)
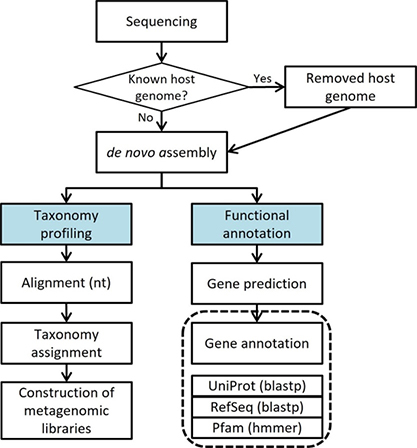
Shotgun Metagenomic Sequencing
- Workflow
-
Bioinformatics
-
Standard Analysis
Remove host sequences
metagenome assembled genomes
Prediction genomic region
Gene annotation
Taxonomy assign
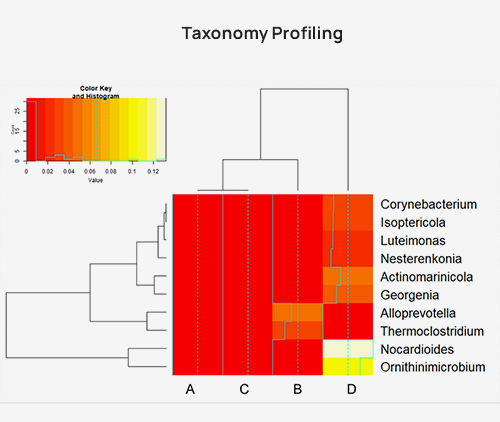
-
Advanced Analysis
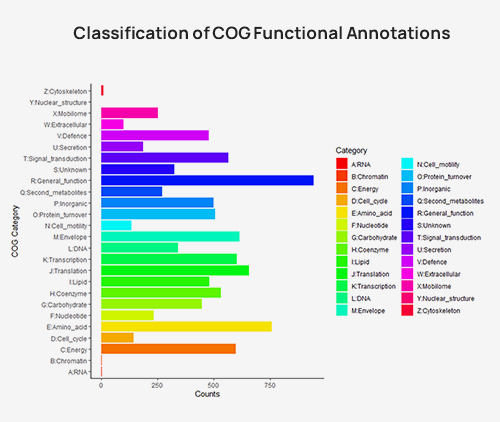
Classification of COG functional annotations
No content
No content
No content
No content
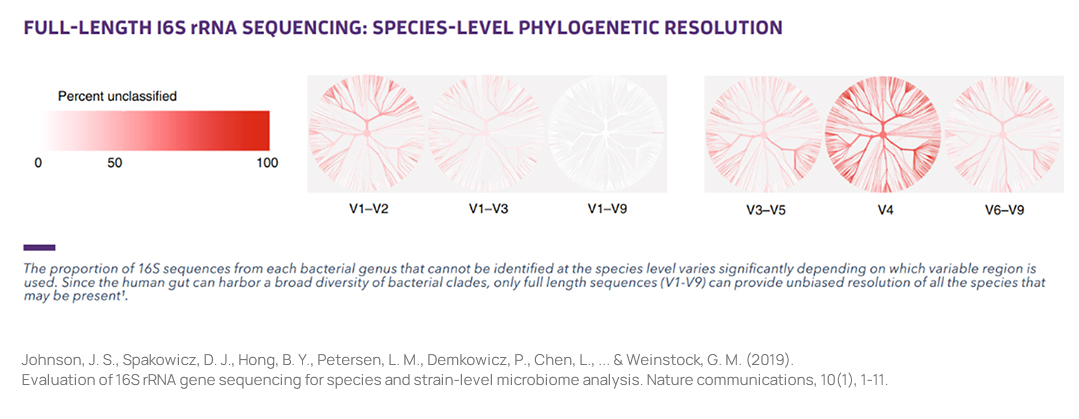
Full Length 16S rRNASequencing
- Workflow
-